Function documentation and dependencies
R package development workshop
June 26, 2024
Overview
- Documenting functions with roxygen2
- NAMESPACE: exporting functions
- NAMESPACE: importing functions
Function documentation with roxygen2
roxygen2
The roxygen2 package generates documentation from specially formatted comments, that we write above the function code, e.g.
#'is a roxygen comment.@paramis a roxygen tag.The
@paramtag takes an argument: the name of the parameterThe remaining text (until the next tag in the file) is the documentation relevant to the tag.
Common tags
There are four tags you’ll use for most functions:
| Tag | Purpose |
|---|---|
| @param arg | Describe inputs |
| @examples | Show how the function works |
| @return | Describe the return value (not needed if NULL) |
| @export | Add this tag if the function should be user-visible |
Usual RStudio shortcuts work in the @examples section, allowing you to run code interactively.
The description block
The roxygen comment should start with a description block.
- First sentence is the title.
- Next paragraph is the description.
- Everything else is the details (optional).
#' Title in Title Case of up to 65 Characters
#'
#' Mandatory description of what the function does.
#' Should be a short paragraph of a few lines only.
#'
#' The details section is optional and may be several paragraphs. It can even
#' contain sub-sections (not illustrated here).RStudio helps you get started
Put your cursor inside a function, then select ‘Insert Roxygen Skeleton’ from the Code menu.
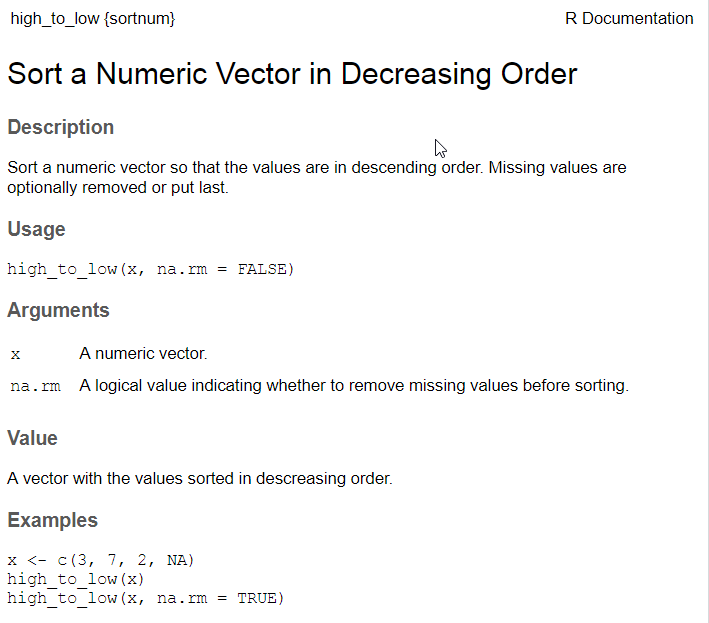
Example roxygen documentation
#' Sort a Numeric Vector in Decreasing Order
#'
#' Sort a numeric vector so that the values are in deceasing order.
#' Missing values are optionally removed or put last.
#'
#' @param x A numeric vector.
#' @param na.rm A logical value indicating whether to remove missing values
#' before sorting.
#' @return A vector with the values sorted in descreasing order.
#' @export
#'
#' @examples
#' x <- c(3, 7, 2, NA)
#' high_to_low(x)
#' high_to_low(x, na.rm = TRUE)R documentation file
roxygen2 converts the roxygen block to an .Rd file in the /man directory
% Generated by roxygen2: do not edit by hand
% Please edit documentation in R/high_to_low.R
\name{high_to_low}
\alias{high_to_low}
\title{Sort a Numeric Vector in Decreasing Order}
\usage{
high_to_low(x, na.rm = FALSE)
}
\arguments{
\item{x}{A numeric vector.}
\item{na.rm}{A logical value indicating whether to remove missing values
before sorting.}
}
\value{
...HTML file
When the package is installed, the .Rd is converted by R to HTML on demand
Regular documentation workflow
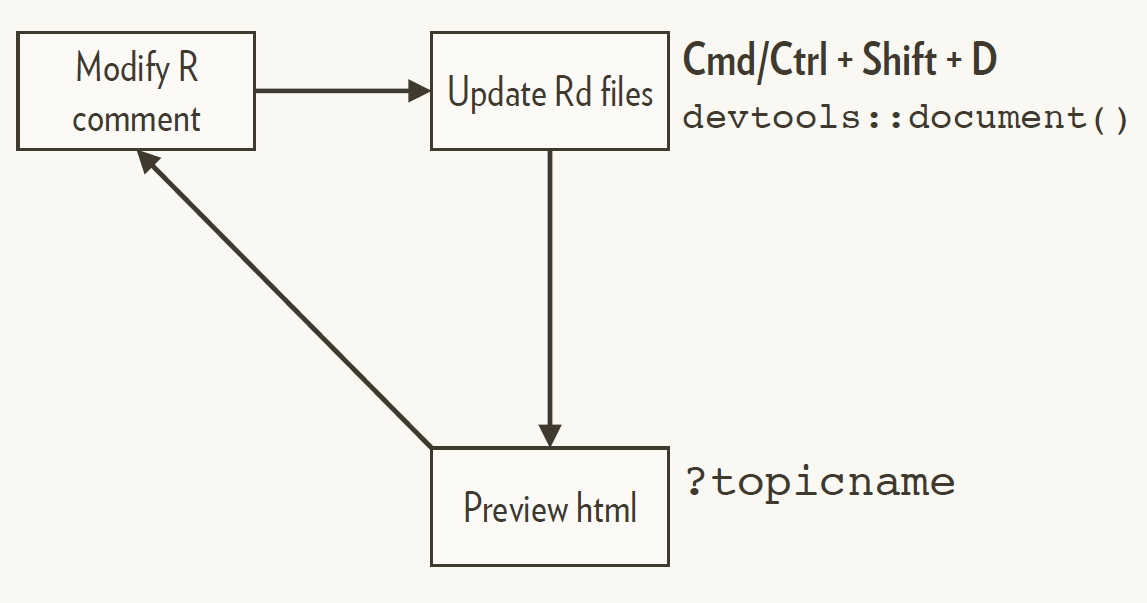
You must have loaded the package with load_all() at least once.
NAMESPACE: exports
A namespace splits functions into two classes
| Internal | External |
|---|---|
| Only for use within package | For use by others |
| Documentation optional | Must be documented |
| Easily changed | Changing will break other people’s code |
Default NAMESPACE
- It is best to export functions explicitly
- The
NAMESPACEfile as created byusethis::create_package()does not export anything by default.
Warning
A package created from the RStudio menus via File > New Project > New Directory > R Package creates a NAMESPACE that exports everything by default, with exportPattern("^[[:alpha:]]+")
This is a good reason not to do this: always call usethis::create_package() to create a package.
For similar reasons, also avoid package.skeleton().
Exporting functions
When we call devtools::document(), an export() directive will be added to NAMESPACE for each function that has an #' @export comment.
# Generated by roxygen2: do not edit by hand
export(fun1)What to export
Only export functions that you want your package users to use, i.e. those that are relevant to the purpose of the package.
Don’t export internal helpers, e.g.
# Defaults for NULL values
`%||%` <- function(a, b) if (is.null(a)) b else a
# Remove NULLs from a list
compact <- function(x) {
x[!vapply(x, is.null, logical(1))]
}Note
Using the ‘Insert Roxygen Skeleton’ option adds an @export tag.
Your turn
For the animal_sounds function:
- Insert a Roxygen skeleton using the RStudio helper.
- Create a draft documentation file with
devtools::document()orCmd/Ctrl + Shift + D. - Click on “Diff” in the Git pane and view the changes that have been made.
- Preview the HTML help with
?animal_sounds. - Fill in the Roxygen skeleton for
animal_sounds(), recreating the documentation file and previewing the HTML help to view your updates. - When you have finished editing, run
devtools::document()to ensure the.Rdfile is in sync. Make a git commit with your updatedR/animal_sounds.Rfile, the updated NAMESPACE, and the newman/animal_sounds.Rdfile.
.Rd Markup
.Rd files recognise LaTeX-like mark-up in most text-based fields, e.g.
#' This is a convenience function that is a wrapper around
#' \code{\link{sort.int}}.Details can be found in the Writing R documentation files section of the Writing R Extensions manual.
Using markdown
Most commonly used mark-up is easier with markdown (and can be mixed with .Rd mark-up).
Text formatting:
**bold**,_italic_,`code`Create links
- To a function in the same package:
[func()] - To a function in a different package:
[pkg::func()] - With different link text, e.g.
[link text][func()]
- To a function in the same package:
For more details, see the (R)Markdown support vignette.
Your turn
- Add some details to the help page for
animal_sounds(), with a link topaste0()and some markdown syntax. - Add a link to a function from a package you don’t have installed (perhaps
basemodels::dummy_classifier()). - Run
devtools::document()and check the link in the help page. What happens? - Run
devtools::check(). Does the link cause problems? - Delete the link to the package you don’t have installed and run
devtools::document()again. - Commit all your changes to the git repo.
Dependencies
Dependencies
Dependencies are other R packages that our package uses. There are three types of dependency:
Imports: required packages, will be installed when our package is installed if they are not already installed.
Suggests: optional packages, e.g. only used for development; only used in documentation. Not installed automatically with our package.
Depends: essentially deprecated for packages, may be used to specify a minimum required version of R (i.e., version of the core packages).
Imported packages
In DESCRIPTION
Imports:
pkgname1
pkgname2Use :: to access functions
Suggested packages
In DESCRIPTION
Suggests:
pkgnameIn package functions or examples, handle the case where pkgname is not available:
use_package()
use_package() will modify the DESCRIPTION and remind you how to use the function.
By default, packages will be added to “Imports”.
NAMESPACE: imports
You might get tired of using :: all the time
Or you might want to use an infix function
You can import functions into the package
devtools::document() will add corresponding import() statements to the NAMESPACE, e.g. import(purr, keep, modify).
Adding formal imports is slightly more efficient than using ::.
Here, the @importFrom tag is placed above the function in which the imported function is used.
Package-level import file
Imports belong to the package, not to individual functions, so alternatively you can recognise this by storing them in a central location, e.g. R/animalsounds-package.R
usethis::use_import_from()
There can be several steps to importing a function. usethis::use_import_from() takes care of all of them.
It will first create the package documentation file R/animalsounds-package.R (if it doesn’t already exist – you will also need to agree to this).
✔ Adding 'purrr' to Imports field in DESCRIPTION
✔ Adding '@importFrom purrr keep', '@importFrom purrr modify' to 'R/animalsounds-package.R'
✔ Writing 'NAMESPACE'
✔ Loading animalsoundsIt may be tempting to import a whole package…
…but it is dangerous
Works today…
… but next year, what if pkg2 adds a fun1 function?
Documenting dependencies
| DESCRIPTION | NAMESPACE |
|---|---|
| Makes package available | Makes function available |
| Mandatory | Optional (can use :: instead) |
use_package() |
use_import_from() |
Example: rlang and cli
Currently we are using stopifnot() for argument validation
We might instead use rlang::is_character() with cli::cli_abort()
Aside: informative messages with cli
cli functions can combine glue interpolation and inline classes to produce informative, nicely-formatted error messages.
In animal_sounds() we can use
This gives the error message
Your turn
- Use
use_package()to addrlangandclitoImports. - Update
animal_sounds()to useis_character()to check the arguments andcli_abortto throw an informative error if necessary, using::to fully qualify the function calls. - Load all and try giving
animal_sounds()invalid inputs for animal and/or sound. - Commit your changes to git.
- Push your commits for this session.
End matter
References
Wickham, H and Bryan, J, R Packages (2nd edn, in progress), https://r-pkgs.org.
R Core Team, Writing R Extensions, https://cran.r-project.org/doc/manuals/r-release/R-exts.html
License
Licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (CC BY-NC-SA 4.0).