Efficient Programming
R Programming
March 18, 2024
Overview
- Memory management
- Benchmarking
- Improving run time
- Parallelisation
- Outlook to package development
Memory management
Overview
Objects created in R are stored in memory. This has the advantage that objects can be accessed faster, but R slows down as the memory fills up. Creating objects also takes time.
Therefore:
- Re-use temporary variables. The allocated storage will be re-used if the vector has the same length.
- Save results for re-use, e.g. index variables
- Don’t save intermediate results unnecessarily – compute on-the-fly
- Remove large objects when no longer needed (with
rm())
Basic data structures
Try to use the simplest data structure for your purpose
- matrices vs. data frames
- character or integer vectors vs. factors
- logical or integer vectors vs. numeric vectors
- unnamed objects vs. named objects
It is especially important to use low-level structures for computation
You can create richer objects as a final step before returning to the user.
Big Data
Modern computers have enough RAM to work with millions of records using standard functions.
Some packages to work more efficiently with big data:
- data.table faster operations on data frames; read/write large CSVs
- dplyr + dbplyr processing of data in databases.
- arrow read/write large CSVs or binary files e.g. Parquet; processing larger-than-memory data with dplyr commands.
- bigmemory, biganalytics faster matrix operations, generalized linear models, kmeans
Growing Objects
Adding to an object in a loop
may force a copy to be made at each iteration, with each copy stored until the loop has completed.
Copy-on-Change
R usually copies an object to make changes to it.
tracemem can be used to trace copies of an object
[1] "<0x122220648>"
[1] "<0x11940ba08>"
[1] "<0x127b9b9c8>"Benchmarking
Benchmarking
There will usually be many ways to write code for a given task. To compare alternatives, we can use benchmark the code.
If the code is more than a single expression, create wrappers for each alternative
bench::mark()
Run the two alternatives with bench::mark(). This function
- Runs alternatives ≥ 1 time; at most enough times to take 0.5s
- Makes sure the two expressions return the same result!
Warning: Some expressions had a GC in every iteration; so filtering is
disabled.# A tibble: 2 × 6
expression min median `itr/sec` mem_alloc `gc/sec`
<bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
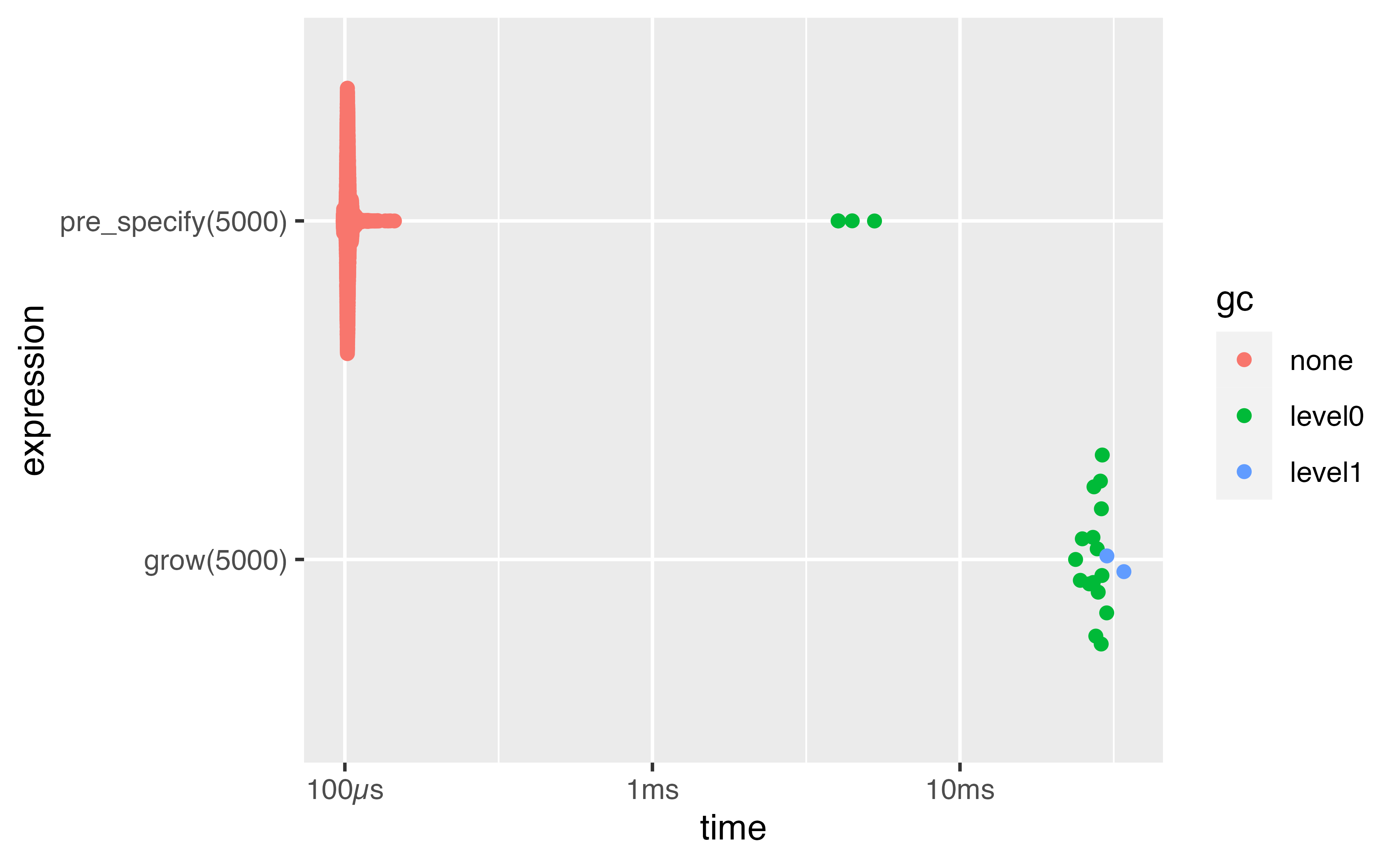
1 grow(5000) 23.8ms 28ms 35.8 95.6MB 65.6
2 pre_specify(5000) 98.7µs 102µs 9362. 55.8KB 8.00GCis the garbage collector which tidies up deleted objectsitr/secis how many times the expression could be run in 1s
Plotting benchmarks
Distribution tends to be right-skewed - focus on the median!
Scaling
Benchmarking can be difficult as the best option can depend on the size of the data, e.g. memory allocation can overshadow run time for small objects.
When thinking about how our code scales to bigger, we need to consider what we mean by “big”
- number of rows or number of columns?
- number of observations or number of factor levels?
bench::press() compares a function over a grid of parameters
bench::press()
bench::press(n = c(10, 100), k = c(10, 1),
bench::mark(gl(n, k, length = 1000)) # `gl` generates factor levels
)Running with:
n k1 10 102 100 103 10 14 100 1# A tibble: 4 × 8
expression n k min median `itr/sec` mem_alloc `gc/sec`
<bch:expr> <dbl> <dbl> <bch:> <bch:> <dbl> <bch:byt> <dbl>
1 gl(n, k, length = 1000) 10 10 3.12µs 3.65µs 245179. 12.4KB 24.5
2 gl(n, k, length = 1000) 100 10 8.86µs 9.43µs 101218. 11.05KB 20.2
3 gl(n, k, length = 1000) 10 1 2.83µs 3.16µs 302419. 3.95KB 30.2
4 gl(n, k, length = 1000) 100 1 5.9µs 6.48µs 146617. 7.53KB 14.7Exercise 1
Suppose we have a matrix of data and a two-level factor
Use bench::mark() to compare the following ways to find the coefficients of a linear model fitted to each row
Improving run time
Faster common operations
- Sorting
- Use
sort(x, partial = 1:10)to get the top 10 - Use
sort(x, decreasing = TRUE)vsrev(sort(x))
- Use
- Generating numeric vectors
seq.int(),seq_along(x),seq_len(n)vsseq()
rep.int()orrep_len(n)vsrep()
which.min(),which.max()vs e.g.which(x == min(x))
anyNA(x)vsany(is.na(x))
For loops
For loops are an intuitive way to write code, but can be very inefficient.
for is a function, : or seq_along is another function, each use of [ is a call to a function, …, so a loop involves many nested function calls.
Try to keep for loops for truly iterative computations or tasks that are fast in any case (optimizing code takes time!)
Otherwise make loops as lean as possible, by pre-computing values that do not need be be computed iteratively.
Vectorization
Vectorization is operating on vectors (or vector-like objects) rather than individual elements.
Many operations in R are vectorized, e.g.
[1] FALSE TRUE FALSE[1] 0.0000000 0.6931472 1.0986123a b
3 6 We do not need to loop through each element!
Recycling
Vectorized functions will recycle shorter vectors to create vectors of the same length
This is particularly useful for single values
and for generating regular patterns
ifelse()
ifelse is a vectorised version of if and else blocks
Recycling is also very useful here
However indexing is more efficient than ifelse
Logical operations
Logical operators such as & and | are vectorized, e.g.
If we only want to compare vectors of length 1 the operators && and || are more efficient as they only compute the RHS if needed
Make sure the vectors are of length 1, otherwise you get an error. This change was introduced in R ≥ 4.3.
Vectorization and Matrices
Vectorizations applies to matices too, not only through matrix algebra
but also vectorized functions
Matrices and recycling: rows
Values are recycled down matrix, which is convenient for row-wise operations
Matrices and recycling: columns
To do the same for columns we would need to explicitly replicate, which is not so efficient.
apply()
apply provides a way to apply a function to every row or column of a matrix
lapply()
lapply applies a given function to each element of a list
sapply() and vapply()
sapply acts similarly to lapply, but tries to simplify the result
It is better to use vapply() in programming as it ensures the returned object is of the expected type (and is slightly faster)
Row/Column-wise Operations
Several functions are available implementing efficient row/column-wise operations, e.g. colMeans(), rowMeans(), colSums(), rowSums(), sweep()
These provide an alternative to iterating though rows and columns in R (the iteration happens in C, which is faster).
The matrixStats provides further “matricised” methods, including medians and standard deviations.
Exercise 2 (h/t Raju Bhakta)
Sampling from 0.3 × N(0, 1) + 0.5 × N(10, 1) + 0.2 × N(3, 0.1):
# Set the random seed and the number of values to sample
set.seed(1); n <- 100000
# Sample the component each value belongs to
component <- sample(1:3, prob = c(0.3, 0.5, 0.2),
size = n, replace = TRUE)
# Sample from the corresponding Normal for each value
x <- numeric(n)
for(i in seq_len(n)){
if (component[i] == 1){
x[i] <- rnorm(1, 0, 1)
} else if (component[i] == 2) {
x[i] <- rnorm(1, 10, 1)
} else {
x[i] <- rnorm(1, 3, sqrt(0.1))
}
}Exercise 2 (continued)
The for loop in the previous code is suitable for vectorization: the iterations are completely independent.
rnorm() is vectorized in the arguments mu and sd, e.g. to simulate a value from the 1st and 3rd component we could write:
[1] -1.371225 3.324096Use this information to replace the for loop, using a single call to rnorm() to simulate n values from the mixture distribution.
Use bench::mark() to compare the two approaches - don’t forget to set the same seed so the simulations are equivalent!
Parallelisation
Parallelisation
Most functions in R run on a single core of your machine. The future.apply package, part of the futureverse, provides parallel versions of all the apply-type functions.
Parallelisation is most straight-forward to implement for embarrassingly parallel problems, such as applying a function to elements of a list.
Example setup
Adapted from https://henrikbengtsson.github.io/course-stanford-futureverse-2023/
Let’s create a slow function:
Parallelising map-reduce calls
Now suppose we have four sets of numeric vectors, in a list, and we want to calculate slow_sum() for each:
We could run lapply() over this, but it takes a while as it handles each list item in turn:
Setting up for parallel processing
The future.apply package comes to the rescue!
The first step is to make a cluster from the available cores.
To parallelise on a local machine, use multisession in plan():
The default number of workers is availableCores().
We’ll also use the tictoc package for timings:
Using future_lapply()
future_lapply() is a drop-in parallel replacement for lapply()
The four slow sums are calculated in about the same time as it takes to calculate one, since they are being calculated simultaneously on separate cores.
Your turn!
The efficient package contains a function to simulate a game of snakes and ladders, returning the number of rolls required to win.
Parallelise the following code:
Use tictoc to compare the run-times. Roughly how large does N have to be for the parallel version to be worth using?
General Principles
Try to use vectorized functions where possible.
Otherwise use the
applyfamily (and parellelise if necessary). Custom functions will often be useful here to pass toapplyetc.Try to keep for loops for true iterative computations or tasks that are fast in any case (optimizing code takes time!)
End matter
References
Good references on optimizing R code:
Wickham, H, Advanced R (2nd edn), Improving performance section, https://adv-r.hadley.nz/perf-improve.html
Gillespie, C and Lovelace, R, Efficient R programming, https://csgillespie.github.io/efficientR/
Tutorials on the Futureverse:
License
Licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (CC BY-NC-SA 4.0).